Screen fewer compounds with diversified scaffolds
Analyze your target and optimize your HTS screening compound library to your target of interest in just a couple of weeks.
Identify off-targets of your compound
De-risk and get control over potential complications at an early stage.
A Knowledge-Based Approach
Our disruptive software technology identifies drug candidates based on the geometry of protein binding sites and their drug-target interactions. This method paves new ways to explore the drug-target space at a ground-breaking precision.
- Predictions based on 3D protein structure & binding affinity data
- Exploits hidden knowledge in vast numbers of protein-ligand complexes found in the Protein Data Bank (PDB)
- Delivers high scaffold diversity
- Off-target identification

Explore PharmAI’s unique accelerated hit finding service
Case study: binders to cGMPdependent 3',5'-cyclic phosphodiesterase (PDE2)
PharmAI’s DiscoveryEngine scored 125 compounds from a library of 2M compounds, spread across the chemical space. After in vitro validation, seven compounds showed binding to PDE2 (at a hit rate of almost 6%) of which three showed affinities in the lower micromolar range.
Deep Learning for Predictions
Our Artificial Intelligence (AI) initiative opens the door to new compound classes. We use classical AI methods with the combination of Deep Learning to predict binding affinity and to improve the quality of our algorithm’s output. Our aim is make the discovery of drugs cheaper and quicker while keeping it at a high precision. In addition, we contineously improve our algorithms by combining classical and semi-supervised deep-learning in a smart manner.
Off-target identification service at an early stage of drug discovery
Case study: Prediction of MAPK14 Off-Targets
Using PharmAI's DiscoveryEngine, 13 off-targets were identified and further validated in the laboratory tests in collaboration with 2bind GmbH. Of the 13 tested proteins, binding was observed for six which corresponds to an extraordinarily high hit rate of 46 percent.
Delivering High-Quality Results, Rapidly
We invested 500 CPU years of pre-computation to ensure a fast delivery of your results just in a few days. Our efficient algorithms make use of this data resource and run order of magnitudes faster than traditional virtual screening.
- Fast turnaround for screening library focus, off-target prediction & scaffold diversification
- Systematic similarity screens across large databases
- Exceptionally high success rates
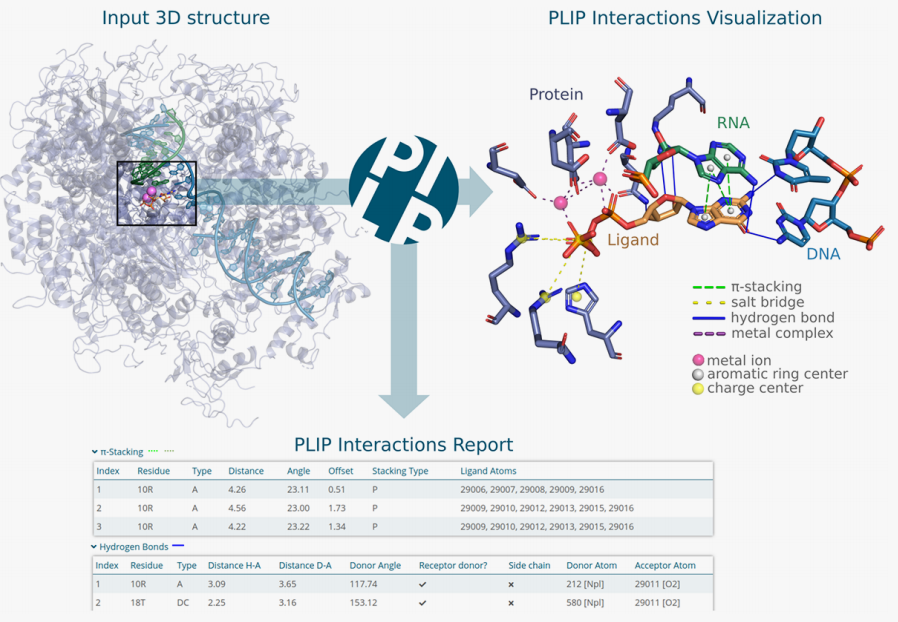
Protein-Ligand Interaction Profiler (PLIP)

PLIP, a command line tool and a web service for a fully automated characterization of non-covalent interactions between proteins and ligands in 3D structures is now available for the community under Apache License at https://github.com/pharmai/plip.
You can also read about and follow the latest developments of PLIP in our blog.
You can also read about and follow the latest developments of PLIP in our blog.
Why should you choose PharmAI?
There are many reasons for choosing us as your partner in your drug discovery journey.
Scaffold Diversification
PharmAI's DiscoveryEngine easily explores new chemical scaffolds for your target.
Accurate
Our fingerprinting technology guarantees an accurate representation of your target of interest.
Fast
We have pre-invested over 500 CPU years of computation time to deliver your results within a few days.
Coverage
We cover over 2.000.000 compounds from multiple vendors and can even use your custom library.